Legume Genome Sequencing Consortium
Shifeng Cheng1, Ashley N. Egan2, Warren Cardinal-McTeague3, Gane Ka-Shu Wong4 & Jeffrey J. Doyle5
1Chinese Academy of Agricultural Sciences, China
2Utah Valley University, USA
3University of British Columbia, Canada
4University of Alberta, Canada
5Cornell University, USA
With the development of genomic sequencing technologies and bioinformatics, the ability to “sequence everything” for an entire genus or even for an entire family, is becoming a reality. Such endeavors enable super-pangenome or extensive comparative evolutionary analyses through dense taxonomic sampling. These advances open new ways to explore diversity in a broad comparative context, leveraging whole-genome sequences to dissect traits of interest in the context of phylogenomics, in order to gain a deeper and more holistic view of the evolution of legumes. We are launching the “Legume Genome Sequencing Consortium”, which aims to generate deep genome sequences of representative species from all major lineages of legumes. This will include broad sampling of representatives of almost all the 770 legume genera, and all major (and many orphan) legume crops and their global diversity panels, as well as trait-based phylogenomics and multi-omics studies. The consortium aims to build an open, inclusive, interactive, and collaborative platform for the community in order to make an impact. We list four of the main components and the existing subprojects that are already under way. Each of these has potential for collaboration with the LPWG and the wider legume community. The fifth section invites brainstorming to propose additional complementary sub-projects, to maximize the value of this initiative and ensure that the consortium is fully inclusive, and creative across the legume community.
-
“Legume Nodulation and NFN Clade Phylogenomics v2.0” Project In this subproject, two parts will be highlighted, aiming to sequence and assemble up to 300 high-quality chromosome-level genomes by combining long-read technologies like PacBio/ONT, Illumina or MGI short-read technologies, and linked read technologies like stLFR or HiC, etc. The first part is to sequence genomes from different legume lineages and or representatives of Fabales lineages which have lost the ability to noduate (including from subfamilies Cercidoideae, Detarioideae, Duparquetioideae, Dialioideae, some Caesalpinioideae, and some Papilionoideae). Species of particular interest will be covered like Nissolia from Dalbergieae, the ADA clade and Swartzieae from Papilionoids, and many taxa phylogenetically encompassing the clade that includes Chamaecrista and mimosoids in the Caesalpinioideae.
The second part is outside the legumes to sequence and assemble the genomes of “comparison pairs” of nodulating and non-nodulating taxa representing the other nine nodulating families across the four orders in the N2-Fixing root Nodule (NFN) clade of angiosperms, which mostly come from the actinorhizal plants. This is a continuation of the v1.0 effort in the EvoNod consortium, launched in 2015 (https://www.science.org/doi/10.1126/science.aat1743). Specifically, the second part will include: 1) outgroups outside the NFN clade (e.g., Malpighiales, and other orders closely related to the NFN clade); 2) actinorhizal nodulators (about 230 species out of 24 genera) and their closely-related non-nodulator relatives, which include Comptonia, Morella, Myrica, Alnus/Betula, Casuarina, Allocasuarina, Ceuthostoma, and Gymnostoma from Fagales; the two subfamilies Datisca and Coriaria from Cucurbitales; Dryas, Purshia, Chamaebatia, Cercocarpus, Elaeagnus, Hippophae, Shepherdia, Trevoa, Retanilla, Ochetophila, Colletia, Discaria, Kentrothamnus, Ceanothus and Parasponia (associated with rhizobia) from Rosales. -
Super-Pangenomics of Legume Crops This effort is aligned with the Global Grain Genomics Research Program (G3RP consortium: https://g3rp.com/) (Cereal + Legume crops). The aim here is to sequence and assemble reference genomes for all known legume crops (grain legumes and other economically important root or tuber crops) and their wild relatives, and to assemble population-based global diversity panels. 100+ new/orphan (economically important) genomes can be sequenced, including various beans, pulses, or other herbaceous legumes. Collaborative possibilities include: 1) building de novo chromosome-level reference-quality genomes for pan-genomic and phylogenomic studies; 2) building population-based global diversity panels and performing whole-genome re-sequencing to build variation maps across numerous accessions of crop species to elucidate crop legume evolution and domestication; 3) collecting germplasm (seeds or other propagules), and further growing the living plants for multi-omics efforts to assemble additional physiological and phenotype data.
-
Legume ENCODE and Gene Mapping Project This Consortium is seeking to collaborate with the LPWG community to develop and combine genomics, phenomics and deep learning particularly in legume model systems to better understand traits and evolution, and the breeding potentials in agriculture: 1) Legume Functional Genomics: building an atlas of gene expression, gene epigenetics and regulation in a multi-omics framework, extending the existing or emerging legume model systems in a legume version of the ‘ENCODE’ project (see pENCODE: https://pubmed.ncbi.nlm.nih.gov/25149370/, or https://www.encodeproject.org/); 2) Gene mapping across the evolutionary tree: mapping or projecting genes, traits and variations of interest across lineages in a phylogenetic context both at the macro-evolutionary and the population levels. This requires a community effort to share, integrate and further develop existing legume datasets from genetics, genomics, and traits, to physiology, ecology, and phenotypes, enabling a joint linkage association study for trait dissection; 3) Deep Learning and legume genomics-based pre-breeding: building a database with a search engine, to integrate genomics and phenomics data for the existing legume model systems, and deploying deep learning with genomics for basic research and pre-breeding. The Pisum sativum [=Lathyrus oleraceus] /(the Mendel pea) diversity panel and model system is already established and provides a good starting point together with several other model systems: Glycine, Lotus, and Medicago.
-
Legume Full-Coverage Phylogenomics Two aspects will be covered: 1) Sequence and assemble 1-2 representative genomes for each of the c. 770 genera of Leguminosae, creating a new comparative framework of “One Thousand Legume Genomes”, and putting the genes/pathway/traits and any evolutionary innovations of interest into this context; 2) Explore legume diversity via collecting RNA-seq samples (transcriptomes) from seeds, flowers and roots/nodules, for each of the sequenced genome/species in Point 1. For more details about these plans, see: https://academic.oup.com/gigascience/article/7/3/giy013/4880447. Funding for sequencing is available and LPWG collaboration is being sought for this initiative from people with specific projects/clades that they could supply material of.
-
Question-Driven and Trait-Based Phylogenomics The aim here is to encourage any ideas that fully utilize these state-of-the-art technologies, like multi-omics, single-cell or spatially-resolved transcriptome, molecular functional genomics and phenotyping technologies and to explore, stimulate and develop additional ideas and question-driven and trait-based collaborative subprojects. We are calling for LPWG involvement in developing and structuring this exciting new initiative to maximize its value and ensure that it meet the needs of the entire global legume community. Please contact the Interim Steering Group if you are passionate, visionary, and motivated to volunteer to serve on the Steering Group, or if you already have particular species, germplasm, expertise, sampling plan, traits or genes of interest, or legume genome sequencing projects that you would like to be included in the context of this initiative. The main tasks and responsibilities of the Steering Group are to provide research direction, set priorities and evaluate research proposals, lobby funding agencies and publicize project progress, coordinate action points and optimize the overall timeline. More brainstorming is needed to get the ball rolling. The sequencing technologies, genomics and bioinformatics are now routinely applied in the Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences (AGIS, CAAS) and the China National GeneBank (CNGB, Shenzhen), the data and analyses will be shared in a timely manner and made publicly available to all participants in the consortium, following the Human Genome Project spirit (“Needed by All, Owned by All, Done by All, and Shared by All”). Seed and plant tissue samples need to be vouchered in recognized herbaria, shared and exchanged via a standard material transfer agreement.
To facilitate planning of these initiatives, Ashley N. Egan (Utah Valley University) has updated the previous list [see Egan & Vatanparast (2019) Australian Systematic Botany 32: 459-483 (https://www.publish.csiro.au/SB/SB19019)] of legume species whose genomes are either fully sequenced or currently being sequenced, whose phylogenetic distribution is illustrated on the following page.
Interim Steering Committee:
Shifeng Cheng, Chinese Academy of Agricultural Sciences, China. chengshifeng@caas.cn
Ashley N. Egan, Utah Valley University, USA. aegan@uvu.edu
Warren Cardinal-McTeague, University of British Columbia, Canada.
Bruno Nevado, Universidade de Lisboa, Portugal. bnevado@fc.ul.pt
Firouzeh Javadi, Kyushu University, Japan. javadisf@yahoo.com
Eric Bishop von Wettberg, University of Vermont, USA. eric.bishop-von-wettberg@uvm.edu
Euan K. James, The James Hutton Institute, UK. euan.james@hutton.ac.uk
Gane Ka-Shu Wong, University of Alberta, Canada. gane@ualberta.ca
Jeffrey J. Doyle, Cornell University, USA. jjd5@cornell.edu

Chamaecrista desvauxii (Collad.) Killip, Caesalpinioideae, photo by Colin Hughes.
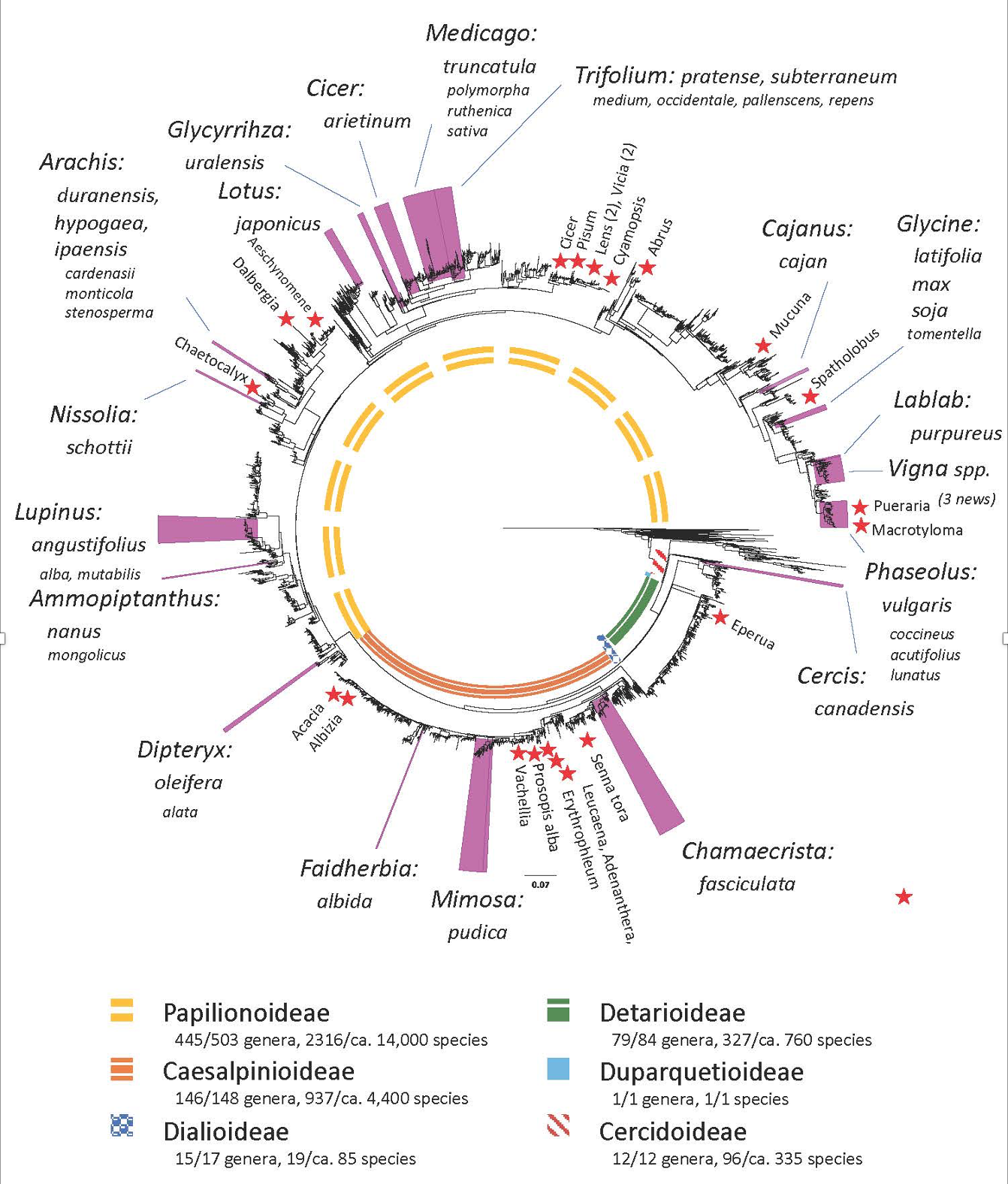 Figure modified from Egan and Vatanparast (2019) with stars and smaller font indicating new additions.
Figure modified from Egan and Vatanparast (2019) with stars and smaller font indicating new additions.