OCCURRENCE DATA WORKING GROUP
Coordinators:
Jens Ringelberg (University of Zürich, Switzerland; moving to University of Edinburgh, UK)
Edeline Gagnon (Technical University Munich, Germany; moving to University of Guelph, Canada)
Joe Miller (Global Biodiversity Information Facility, GBIF)
2022 has been a productive year for legume biogeography research. Multiple research groups have spent a lot of time and energy assembling substantial new quality-controlled occurrence datasets, several of which are (nearly) finished and being used to address various exciting research questions about legume biogeography.
Moabe Fernandes and Toby Pennington (University of Exeter, U.K.) are compiling an occurrence dataset for all legumes in the Americas. They are progressing well, have assembled data for almost 90% of their target species and are already producing extinction risk assessments and mapping areas with high densities of threatened species. Their next step will be to combine their occurrence data with phylogenetic information to investigate patterns of phylogenetic diversity across the Americas.
In her MSc thesis, Charlotte Hagelstam-Renshaw (Université de Montréal), supervised by Anne Bruneau (Université de Montréal) and Warren Cardinal-McTeague (University of British Columbia), has compiled occurrence data and species distribution maps for the 14 genera of subfamily Cercidoideae, as part of a study of biome evolution across the subfamily. In addition, the Bruneau and Cardinal-McTeague labs have also assembled cleaned occurrence data for a majority of the 85 species (17 genera) in subfamily Dialioideae.
Jens Ringelberg, supervised by Colin Hughes (University of Zurich), has assembled and cleaned species-level occurrence data for 93% of the ca. 3,500 species representing all 100 genera in the mimosoid clade (subfamily Caesalpinioideae). Together with Erik Koenen (University of Brussels) and a large number of collaborators, they have used this large new occurrence dataset to assess phylogenetic turnover of mimosoids across the tropics. Their aim was to gain insights into how a single legume clade was able to diversify across all major lowland tropical biomes on every tropical continent and a paper resulting from this work is now in press.
Finally, the upcoming Advances in Legume Systematics, ALS14, Part 2 will feature a synopsis of the 163 genera in subfamily Caesalpinioideae, including detailed distribution maps for all genera based on quality-controlled occurrence data. ALS14 is a highly collaborative project; occurrence data of non-mimosoid Caesalpinioideae have been contributed by Juliana Rando (Universidade Federal do Oeste da Bahia, Brazil), Guilherme Sousa (Universidade Estadual de Campinas, Brazil), Haroldo de Lima (Instituto de Pesquisas Jardim Botânico do Rio de Janeiro, Brazil) and Domingos Cardoso (Universidade Federal do Bahia & Jardim Botânico do Rio de Janeiro, Brazil), and the data have been compiled and cleaned with the help of many legume researchers.
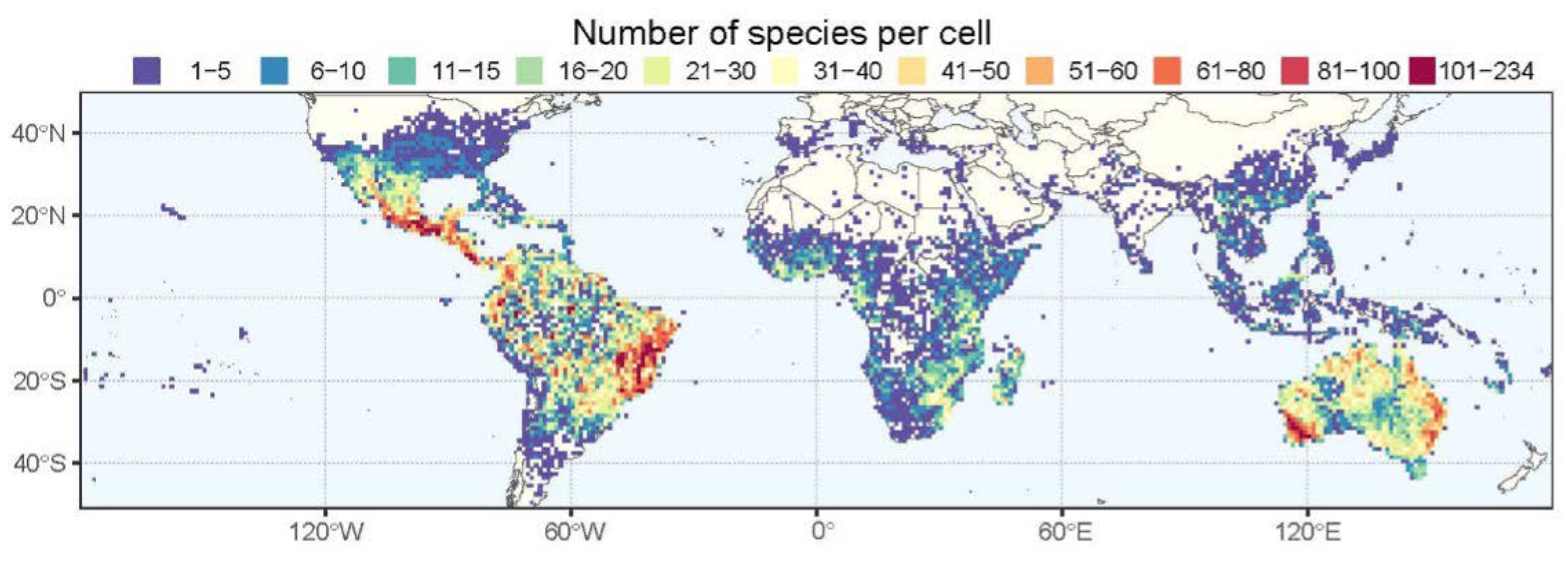 The distribution of Caesalpinioideae species-richness in one-degree cells across the tropics (Advances in Legume Systematics, ALS14, Part 2, in prep.).
The distribution of Caesalpinioideae species-richness in one-degree cells across the tropics (Advances in Legume Systematics, ALS14, Part 2, in prep.).
Publications from all these projects are expected in the near future, meaning that 2023 looks set to be another very exciting year in legume biogeography research.
As a researcher it can sometimes be easy to forget that compiling (occurrence) datasets is not the only thing that matters; equally important is how the resulting datasets are stored and made available. Over the past few years, legume researchers have curated various spatial datasets to address specific research questions. These data are usually published with the manuscript as supplementary data, often deposited in Dryad, Figshare, Zenodo and the likes. A lot of work goes into compiling and cleaning these datasets and their reuse can save other people time. Keep your eyes on the Legume Data Portal, where many of these maps based on cleaned occurrence datasets will be made available, and as we continue to explore options for storing both the maps and data. Ultimately, the curated datasets could be integrated into one larger legume-wide global dataset once any discrepancies in data cleaning protocols are reconciled.
GBIF is also exploring how to make these cleaned, quality-controlled data available. They are exploring a rule-based system that would flag occurrences during indexing in a similar way to a researcher using R scripts for data cleaning. GBIF hopes to have a demonstration ready for the ILC8 conference in Brazil in August. The goal is that the legume community could come up with a set of rules that describe quality controlled data that GBIF can implement. Stay tuned!
Also from GBIF, take a look at the new clustering feature to search for similarities in occurrence data fields across datasets shared with GBIF. Joe Miller has used this feature to identify putative duplicate specimens of Senegalia that were collected in Brazil. Check out Joe’s GBIF data blog post about the patterns of specimen data sharing between Brazilian herbaria and large northern hemisphere herbaria. Please provide feedback, make comments, and share ideas in the discussion on the GBIF community forum.
Finally, the Occurrence Data Working Group needs your help! As an initial step to make cleaned legume occurrence datasets more easily accessible we are assembling a list with links to all existing datasets that are available online alongside who assembled the data and references to relevant papers. Our aim is to create a list that can easily be updated whenever new data become available. If you have a dataset that you would like to be included, please send us a message!